
CFDE 2025 Spring Meeting
March 25-26, 2025
Bethesda Marriott
5151 Pooks Hill Road
Bethesda, Maryland 20814
*Submissions for Abstracts and/or Demos has closed.*
**Registration has closed. If you need to register please contact Swathi Thaker.**
Common Fund Data Ecosystem (CFDE) will host a two-day, in-person program meeting in Bethesda, MD on March 25th and 26th. This will be an opportunity for CFDE members to come together to share projects and accomplishments across the Consortium. There will be opportunities for posters and demos as well!
Program Committee and Leadership

Jyl Boline, PhD
Jyl Boline, PhD
Founder, Informed Minds

Jake Chen, PhD
Jake Chen, PhD
University of Alabama at Birmingham
Professor, Dept. of Biomedical Informatics
& Data Science
Director, Systems AI Pharmacology Center (SPARC)

Christy Kano, PhD
Christy Kano, PhD
National Institutes of Health (NIH)
CFDE Coordinator and Project Manager
Office of Strategic CoordinationDivision of Program Coordination, Planning, and Strategic Initiatives
Office of the Director

Chris Kinsinger, PhD
Chris Kinsinger, PhD
National Institutes of Health (NIH)
Assistant Director for Catalytic Data Resources
Office of Strategic Coordination
Division of Program Coordination, Planning, and Strategic Initiatives
Office of the Director

Sahana Kukke, PhD
Sahana Kukke, PhD
National Institutes of Health (NIH)
CFDE Program Officer
Program Leader
Office of Strategic Coordination
Division of Program Coordination, Planning, and Strategic Initiatives
Office of the Director

Jennifer Burnette, MPH
Jennifer Burnette, MPH
ORAU
Program Manager

Alissa Dillman, PhD
Alissa Dillman, PhD
BioData Sage
Founder and CEO

Avi Ma’ayan, PhD
Avi Ma’ayan, PhD
Icahn School of Medicine at Mount Sinai
Mount Sinai Endowed Professor in Bioinformatics
Professor, Department of Pharmacological Sciences
Director, Mount Sinai Center for Bioinformatics

Mano Maurya, PhD
Mano Maurya, PhD
University of California San Diego
Senior Scientist
Department of Bioengineering

Rahi Navelkar
Rahi Navelkar
Harvard Medical School
Bioinformatics Associate and Data Wrangler

George Papanicolaou, PhD
George Papanicolaou, PhD
National Institutes of Health (NIH)
CFDE Program Officer
Program Manager
Office of Strategic Coordination
Division of Program Coordination, Planning, and Strategic Initiatives
Office of the Director

Peipei Ping, PhD
Peipei Ping, PhD
University of California, Los Angeles
Professor, Dept. of Professor of Physiology, Medicine/Cardiology, and Biomedical Informatics

Jonathan Silverstein, PhD
Jonathan Silverstein, PhD
University of Pittsburgh
Chief Research Informatics Officer, Health Sciences and Institute for Precision Medicine (IPM)
Professor, Department of Biomedical Informatics (DBMI), School of Medicine

Shankar Subramanium, PhD
Shankar Subramanium, PhD
University of California San Diego
Distinguished Professor of Bioengineering, Bioinformatics and Systems Biology, Computer Science & Engineering, Cellular & Molecular Medicine and Nano Engineering

Swathi Thaker, PhD
Swathi Thaker, PhD
University of Alabama at Birmingham
Program Operations Manager
Systems AI Pharmacology Center (SPARC)
Agenda
–
| Time (EST) | Session Title & Presenter | Location |
|---|---|---|
| 7:30 – 8:45 | Registration & Breakfast | Grand Ballroom |
| 8:45 – 9:00 | Welcome by Jake Chen | Grand Ballroom |
| 9:00 – 9:15 | NIH Opening Remarks by Chris Kinsinger | Grand Ballroom |
| 9:15 – 10:45 | Session 1: CFDE Center Updates DRC, KC, ICC, Cloud Computing, Training Moderator: Christy Kano | Grand Ballroom |
| 10:45 – 11:00 | Coffee Break | |
| 11:00 – 12:30 | Session 2: Innovative Tools to Analyze Common Fund Data Moderator: Deanne Taylor | Grand Ballroom |
| 12:30 – 1:30 | Lunch | On-Site |
Breakout: Tech Showcase (Descriptions)
| Time (EST) | Track 3A | Track 3B | Track 3C |
|---|---|---|---|
| Location | Bethesda Potomac | Rockville/Chevy Chase | Salon E |
| 1:30 – 1:50 | Virtual Reality Enables Multiscale Exploration of the Human Reference Atlas | Community Visualization Hub: Current Features and Roadmap | Digital Scavenger Hunt: Come explore datasets, tools, and portals within the CFDE in an interactive and engaging way. Participants should bring a laptop, Chromebook or iPad with them to this event. |
| 1:50 – 2:10 | GeneSetCart: Assembling, Augmenting, Combining, Visualizing, and Analyzing CFDE Gene Sets | Playbook Workflow Builder: Interactive Construction of Bioinformatics Workflows from Common Fund Supported Building Blocks | Digital Scavenger Hunt |
| 2:10-2:30 | CFDE Talent Knowledge Graph | Graph Query Interface for C2M2 Data Search and Discovery | Digital Scavenger Hunt |
| Time (EST) | Session Title & Presenter | Location |
|---|---|---|
| 2:30 – 2:45 | Coffee Break | |
| 2:45 – 3:45 | Session 4 (Parallel a/b/c) Junior Scholar Lightning Talks Moderator: Allissa Dillman | Grand Ballroom |
| 3:45 – 5:00 | Session 5. Poster Presentation & Judging | Grand Ballroom Foyer |
| 5:00 – 6:00 | Networking Reception | |
| END OF DAY 1 |
| Time (EST) | Session Title & Presenter | Location |
|---|---|---|
| 7:30 – 8:15 | Breakfast | |
| 8:15 – 8:30 | Day 2 Agenda Overview by Jake Chen | Grand Ballroom |
Breakout: Working Groups
| Time (EST) | Track 5A | Track 5B | Track 5C | Track 5D |
|---|---|---|---|---|
| Location | Bethesda Potomac | Rockville/Chevy Chase | Salon E | Grand Ballroom |
| Topic / Host | Ontology Mano Maurya and Srini Ramachandran | Knowledge Graph Deanne Taylor and Johnathan Silverstein | Trainers Allissa Dillman and Jennifer Burnette | Outreach Noel Burtt and MacKenzie Brandes |
| 8:30 – 9:00 | Overview of C2M2 and metadata submission (ends at 9:15) | Updates on Data Distillery (current state of DDKG) | Summer Mentorship Program (ends at 9:15) | Charter Finalization |
| 9:00 – 9:30 | (Begins at 9:15) Addressing pending topics/questions from OWG meetings | Community Updates | Landscape Analysis Results and Collaboratively working to address Recommendations | Outreach Tools and Resources |
| 9:30-10:00 | Technologies and standards; LLMs and KGs | None | Future Events |
10:00 – 10:15 – Coffee Break
Breakout: Partnership & Science
| Track 6A | Track 6B | Track 6C | |
|---|---|---|---|
| Location | Bethesda Potomac | Rockville/Chevy Chase | Salon E |
| 10:15 – 11:00 | Biomarker (Closed Session) | Gene Set Curation (Open to all) | Community Visualization Hub: Collaborative Data Visualization Platform for Genomics and Single-cell data (Open to all) Topic: End User Perspectives – Using the Hub for research |
| 11:00 – 11:45 | Metabotyping (Closed Session) | Gene Set Curation (Open to all) | Community Visualization Hub: Collaborative Data Visualization Platform for Genomics and Single-cell data (Open to all) Topic: Tool Builder Perspectives – Incorporating the Hub in your software |
| Time (EST) | Session Title & Presenter | Location |
|---|---|---|
| 11:45 – 1:00 | Lunch | On-site |
| ↓ | Interactive Panels: Lessons & Future | ↓ |
| 1:00 – 1:30 | Bridging Communities: Integrating CFDE with External Data Networks Moderator: Jeffrey Grethe | Grand Ballroom |
| 1:30 – 2:00 | Sustaining the CFDE Ecosystem Moderators: Peipei Ping and Wei Wang | Grand Ballroom |
| 2:00 – 2:30 | Strategies to Grow the CFDE User Base Moderators: Bernard de Bono and Noel Burtt | Grand Ballroom |
| 2:30 – 3:00 | Townhall | Grand Ballroom |
| 3:00 – 3:10 | Conclusion Moderator: NIH | Grand Ballroom |
| END OF DAY 2 |
1. Leveraging Large-Scale Electronic Healthcare Records with Oracle Real World Data to Illuminate Precision Molecular Biomarkers: Troponin and PSA Use Case Development
Vincent T. Metzger, Cristian G. Bologa, Noah G. Reboul, Christophe G. Lambert and Jeremy J. Yang, University of New Mexico
2. KG2ML: Integrating Knowledge Graphs and Positive Unlabeled Learning for Identifying Disease-Associated Genes with Case Studies for 12 Diseases
Praveen Kumar, University of New Mexico
3. Spatially varying cell-specific gene regulation network inference
Yurui Li, UIUC; Jin Chen, Cleveland Clinic; Haohan Wang, UIUC
4. KG-UI: Interactive Web Interface for Biomedical Knowledge Graph Applications
John Erol Evangelista, Anna I. Byrd, Andrew Lutsky, Ho-Young Chung, Alexander Lachmann, Giacomo B. Marino, Sherry L. Jenkins, and Avi Ma’ayan, Mount Sinai Center for Bioinformatics
5. ChEA-KG: Human Transcription Factor Regulatory Network with a Knowledge Graph Interactive User Interface
Anna I. Byrd, John Erol Evangelista, Alexander Lachmann, Ho-Young Chung, Sherry L. Jenkins and Avi Ma’ayan, Mount Sinai Center for Bioinformatics
6. Identifying exercise-mimetic drugs using LINCS and MoTrPAC data
Pauline Brochet, David Jimenez-Morales, Malene E Lindholm, Matthew T Wheeler and Daniel H. Katz, Stanford University
7. Designing and Building a Simple, Scalable, and Extensible Data Ingestion Pipeline
Joseph Okonda, Qiuyue Liu, Jared Nedzel and Kristin Ardlie, Broad Institute
8. L2S2: Chemical Perturbation and CRISPR KO LINCS L1000 Signature Search Engine
Giacomo B. Marino, John E. Evangelista, Daniel J. B. Clarke and Avi Ma’ayan, Mount Sinai Center for Bioinformatics
9. Advancing Training in the Common Fund Data Ecosystem: Insights from a Landscape Analysis
DeBran Tarver, Oak Ridge Associated Universities (ORAU); Kristi Sadowski, Oak Ridge Associated Universities (ORAU); Jennifer Burnette, Oak Ridge Associated Universities (ORAU); Allissa Dillman, BioData Sage LLC; LaFrancis Gibson, Oak Ridge Associated Universities (ORAU); David Burns, Oak Ridge Associated Universities (ORAU); Diane Krause, Oak Ridge Associated Universities (ORAU)
10. BiomarkerKB: A Comprehensive Biomarker Knowledgebase
Daniall Masood, The George Washington School of Medicine
11. The CFDE Workbench: Integrating Metadata and Processed Data from Common Fund Programs
John Erol Evangelista, Mount Sinai Center for Bioinformatics; Daniel J.B. Clarke, Mount Sinai Center for Bioinformatics; Zhuorui Xie, Mount Sinai Center for Bioinformatics; Stephanie Olaiya, Mount Sinai Center for Bioinformatics; Heesu Kim, Mount Sinai Center for Bioinformatics; Giacomo B. Marino, Mount Sinai Center for Bioinformatics; Anna Byrd, Mount Sinai Center for Bioinformatics; Shivaramakrishna Srinivasan, University of California San Diego; Sumana Srinivasan, University of California San Diego; Mano R. Maurya, University of California San Diego; Sherry L. Jenkins, Mount Sinai Center for Bioinformatics; Andrew D. Lutsky, Mount Sinai Center for Bioinformatics; Lucas Sasaya, Mount Sinai Center for Bioinformatics; Alexander Lachmann, Mount Sinai Center for Bioinformatics; Nasheath Ahmed, Mount Sinai Center for Bioinformatics; Ido Diamant, Mount Sinai Center for Bioinformatics; Ethan Sanchez, University of California San Diego; Srinivasan Ramachandran, University of California San Diego; Shankar Subramaniam, University of California San Diego; Avi Ma’ayan, Mount Sinai Center for Bioinformatics
12. Cultivating Tomorrow’s Scientists: Insights from the 2024 CFDE-GlyGen Internship and 2025 Internship plans
Jeet Vora, The George Washington University; Melody Perlman-Porterfield, University of Georgia; Rene Ranzinger, The George Washington University; Michael Tiemeyer, University of Georgia; Raja Mazumder, The George Washington University
13. Preparing and Serving Common Supported Highly Processed Datasets in the Croissant Metadata Standard
Ido Diamant, Daniel J. B. Clarke and Avi Ma’ayan, Mount Sinai Center for Bioinformatics
14. The CFDE Data Distillery Project
Benjamin J. Stear, The Children’s Hospital of Philadelphia; Taha Mohseni Ahooyi, The Children’s Hospital of Philadelphia; J. Alan Simmons, The University of Pittsburgh; Christopher M. Nemarich, The Children’s Hospital of Philadelphia; Jonathan C. Silverstein, The University of Pittsburgh; Deanne M. Taylor, The Children’s Hospital of Philadelphia & University of Pennsylvania
15. GeneSight: Advancing Insights into Complex Genetic Traits through CFDE-Enriched Knowledge Graphs
Cheng-Han Chung, Chris Bizon, Corbin Jones, Jason Reilly, Matthew Niederhuber and Evan Morris, University of North Carolina Chapel Hill
16. Inferring tissue aging clocks from blood transcriptomics
Minja Belic, Buck Institute for Research on Aging
17. Defining the Human Metabotype through Genetic, Diet, Environmental Exposure and Exercise Perturbations
Sara Rahiminejad, University of California, San Diego; Mano R. Maurya, University of California, San Diego; David Jimenez-Morales, Stanford University; Daniel H Katz, Stanford University; Jimmy Zhen, Stanford University; Wimal Pathmasiri, University of North Carolina Chapel Hill; Blake Rushing, University of North Carolina Chapel Hill; Susan McRitchie, University of North Carolina Chapel Hill; Sumana Srinivasan, University of California, San Diego; Srinivasan Ramachandran, University of California, San Diego; Eoin Fahy, University of California, San Diego; Vishal Midya, Icahn School of Medicine at Mount Sinai; Lauren Petrick, Icahn School of Medicine at Mount Sinai; Susan L. Teitelbaum, Icahn School of Medicine at Mount Sinai; Susan Sumner, University of North Carolina Chapel Hill; Matthew T. Wheeler, Stanford University; Shankar Subramaniam, University of California, San Diego
18. Characterizing the Immune Landscape of Pediatric and Adult Cancers
Tianyuzhou Liang, Li-Ju Wang, Yi-Nan Gong, and Yu-Chiao Chiu, University of Pittsburgh School of Medicine
19. The EDRN Knowledge Environment for Cancer Biomarker Research
Daniel Crichton, Heather Kincaid, Alphan Altinok, NASA Jet Propulsion Laboratory and Kristen Anton, University of North Carolina Chapel Hill
20. Expanding the CFDE Data Distillery Knowledge Graph for Childhood Cancer and Birth Defects Research: a CFDE U24 project
Benjamin J. Stear, The Children’s Hospital of Philadelphia; Taha Mohseni Ahooyi, The Children’s Hospital of Philadelphia; Dave D. Hill, The Children’s Hospital of Philadelphia; Ryan Corbett, Indiana University; J. Alan Simmons, University of Pennsylvania; Rebecca Kaufman, The Children’s Hospital of Philadelphia; Adam Resnick, The Children’s Hospital of Philadelphia & Indiana University; Sarah Tasian, The Children’s Hospital of Philadelphia & University of Pennsylvania; Kristina Cole, The Children’s Hospital of Philadelphia; Jonathan Silverstein, University of Pennsylvania; Elizabeth Goldmuntz, The Children’s Hospital of Philadelphia & University of Pennsylvania; Jo Lynne Rokita, Indiana University; Sharon Diskin, The Children’s Hospital of Philadelphia & University of Pennsylvania; Deanne M. Taylor, The Children’s Hospital of Philadelphia
21. Building the Developmental Human Reference Atlas (dHRA): A CFDE-Integrated Framework for Pediatric Anatomical and Genomic Data
Taha Mohseni Ahooyi, The Children’s Hospital of Philadelphia; David Biko, The Children’s Hospital of Philadelphia & University of Pennsylvania; Axel Bolin, Indiana University; Andreas Bueckle, Indiana University; Bruce W. Herr II, Indiana University; Shivaram Karandikar , The Children’s Hospital of Philadelphia; Rebecca Linn, The Children’s Hospital of Philadelphia & Indiana University; Elizabeth Maier, Indiana University; Nancy Ruschman, Indiana University; Jakob Seidlitz, The Children’s Hospital of Philadelphia & University of Pennsylvania; Shiping Zhang, The Children’s Hospital of Philadelphia; Katy Börner, Indiana University; Deanne M. Taylor, The Children’s Hospital of Philadelphia & University of Pennsylvania
22. Gene Set Foundation Model Applied to Augment Gene Sets Created from Common Fund Supported Resources
Daniel J. B. Clarke and Avi Ma’ayan, Mount Sinai Center for Bioinformatics
23. Crosscut Metadata Model (C2M2) Parsing for Enhanced Search on the CFDE Data Portal
Shivaramakrishna Srinivasan, University of California, San Diego & CFDE Data Resource Center (DRC); Ethan Sanchez, University of California, San Diego & CFDE Data Resource Center (DRC); Sumana Srinivasan, University of California, San Diego & CFDE Data Resource Center (DRC); Mano R. Maurya, University of California, San Diego & CFDE Data Resource Center (DRC); Srinivasan Ramachandran, University of California, San Diego & CFDE Data Resource Center (DRC); Daniel J.B. Clarke, CFDE Data Resource Center (DRC) & Mount Sinai Center for Bioinformatics; Avi Ma’ayan, CFDE Data Resource Center (DRC) & Mount Sinai Center for Bioinformatics; Shankar Subramaniam, University of California, San Diego & CFDE Data Resource Center (DRC) & San Diego Supercomputer Center
24. Building a Bioinformatics Ecosystem for Large-Scale Multi-Omics Research: Lessons from MoTrPAC
David Jimenez-Morales, Stanford University School of Medicine; David Amar, Stanford University School of Medicine; Malene E. Lindholm, Stanford University School of Medicine; Mihir Samdarshi, Stanford University School of Medicine; Archana N Raja, Stanford University School of Medicine; Jimmy Zhen, Stanford University School of Medicine; Jiye Yu, Stanford University School of Medicine; Sam Montalvo, Stanford University School of Medicine; Daniel H Katz, Stanford University School of Medicine; Ashley Xia, National Institute of Diabetes and Digestive and Kidney Diseases; Euan A Ashley, Stanford University School of Medicine; Matthew T Wheeler, Stanford University School of Medicine
25. Detection of Aberrant Expression, Rare Variant, and Splicing Outliers in Rare Disease Patients – CFDE Transcriptome Variability Partnership
Archana Natarajan Raja, Stanford University School of Medicine; Alex Miller, Stanford University School of Medicine; Abhishek Choudhary, Broad Institute; Lisa Anderson, Broad Institute; Matthew Wheeler, Stanford University School of Medicine; Stephen B. Montgomery, Stanford University School of Medicine, Kristin Ardlie, Broad Institute
26. Probing the Breadth of the Molecular Response to Exercise using Common Funds Metabolomics Data
Christopher Patsalis, Abraham Raskind, Alla Karnovsky, Charles F. Burant and Charles R. Evans, University of Michigan
27. STRIDE Principles for AI-Ready and Sustainable Datasets
Catilin Ree, CFDE ICC-SC, UCLA Department of Physiology; Steven Swee, CFDE ICC-SC, UCLA Department of Physiology, Medical Informatics/Bioinformatics IDP; Erika Yilin Zheng, CFDE ICC-SC, UCLA Department of Physiology; Miriam Ojeda, CFDE ICC-SC, UCLA Department of Physiology; Jack Rincon, CFDE ICC-SC, Bridge2AI Center TRM & Integrated Cardiovascular Data Science Training Program, UCLA Department of Physiology; Irsyad Adam, CFDE ICC-SC, UCLA, Medical Informatics/Bioinformatics IDP; Dean Wang, CFDE ICC-SC, Bridge2AI Center TRM & Integrated Cardiovascular Data Science Training Program, UCLA Department of Physiology; Wei Wang, CFDE ICC-SC, Bridge2AI Center TRM & Integrated Cardiovascular Data Science Training Program, Medical Informatics/Bioinformatics IDP, Department of Computer Science, UCLA; Peipei Ping, CFDE ICC-SC, Bridge2AI Center TRM & Integrated Cardiovascular Data Science Training Program, UCLA Department of Physiology, Medical Informatics/Bioinformatics IDP, Department of Medicine, Cardiology Division, UCLA
28. Preliminary Analysis of Existing CFDE Programs and Suggestions for their Future Data Hosts
Ethan Ji, CFDE ICC-SC, Bridge2AI Center TRM & Integrated Cardiovascular Data Science Training Program, UCLA Department of Physiology; Catilin Ree, CFDE ICC-SC, UCLA Department of Physiology; Namuna Pandy, CFDE ICC-SC, UCLA Department of Physiology; Miriam Ojeda, CFDE ICC-SC, UCLA Department of Physiology; Jack Rincon, CFDE ICC-SC, Bridge2AI Center TRM & Integrated Cardiovascular Data Science Training Program, UCLA Department of Physiology; Dean Wang, CFDE ICC-SC, Bridge2AI Center TRM & Integrated Cardiovascular Data Science Training Program, UCLA Department of Physiology; Wei Wang, CFDE ICC-SC, Bridge2AI Center TRM & Integrated Cardiovascular Data Science Training Program, Medical Informatics/Bioinformatics IDP, Department of Computer Science, UCLA; Peipei Ping, CFDE ICC-SC, Bridge2AI Center TRM & Integrated Cardiovascular Data Science Training Program, UCLA Department of Physiology, Medical Informatics/Bioinformatics IDP, Department of Medicine, Cardiology Division, UCLA
Tuesday, March 25
| 7:30 – 8:45 (Grand Ballroom) Registration & Breakfast |
| 8:45 – 9:00 (Grand Ballroom) Welcome by Jake Chen |
| 8:40 – 9:00 (Grand Ballroom) NIH Opening Remarks by Chris Kinsinger |
| 9:15 – 10:45 (Grand Ballroom) Session 1: CFDE Center Updates DRC, KC, ICC, Training, Cloud Computing Moderator: Christy Kano |
| 10:45 – 11:00 Coffee Break |
| 11:00 – 12:30 (Grand Ballroom) Session 2: Innovative Tools to Analyze Common Fund Data Moderator: Deanne Taylor |
| 12:30 – 1:30 Lunch |
| Breakout: Tech Showcase |
|---|
| 1:30 – 1:50 Track 3A (Bethesda Potomac) Virtual Reality Enables Multiscale Exploration of the Human Reference Atlas Track 3B (Rockville/Chevy Chase) Community Visualization Hub: Current Features and Roadmap Track 3A (Salon E) Digital Scavenger Hunt: Come explore datasets, tools, and portals within the CFDE in an interactive and engaging way. Participants should bring a laptop, Chromebook or iPad with them to this event. |
| 1:50 – 2:10 Track 3A (Bethesda Potomac) GeneSetCart: Assembling, Augmenting, Combining, Visualizing, and Analyzing CFDE Gene Sets Track 3B (Rockville/Chevy Chase) Community Visualization Hub: Current Features and Roadmap Track 3A (Salon E) Digital Scavenger Hunt: Come explore datasets, tools, and portals within the CFDE in an interactive and engaging way. Participants should bring a laptop, Chromebook or iPad with them to this event. |
| 2:10-2:30 Track 3A (Bethesda Potomac) CFDE Talent Knowledge Graph Track 3B (Rockville/Chevy Chase) Graph Query Interface for C2M2 Data Discovery Track 3A (Salon E) Digital Scavenger Hunt: Come explore datasets, tools, and portals within the CFDE in an interactive and engaging way. Participants should bring a laptop, Chromebook or iPad with them to this event. |
| 2:30 – 2:45 (Grand Ballroom) Coffee Break |
| 2:45 – 3:45 (Grand Ballroom) Session 4 (Parallel a/b/c) Junior Scholar Lightning Talks Moderator: Allissa Dillman |
| 3:45 – 5:00 (Grand Ballroom Foyer) Session 5. Poster Presentation & Judging |
| 5:00 – 6:00 Networking Reception |
| End of Day 1 |
Wednesday, March 26
| 7:30 – 8:15 Breakfast |
| 8:15 – 8:30 (Grand Ballroom) Day 2 Agenda Overview by Jake Chen |
| Breakout: Working Groups |
|---|
| 8:30 – 9:00 Track 5A (Bethesda Potomac) – Ontology Working Group Moderators: Mano Maurya and Srini Ramachandran Topic: Overview of C2M2 and metadata submission (8:30-9:15am) Track 5B (Rockville/Chevy Chase) – Knowledge Graph Working Group Moderators: Deanne Taylor and Jonathan Silverstein Topic: Updates on Data Distillery (current state of DDKG) Track 5C (Salon E) – Trainers Working Group Moderators: Allissa Dillman and Jennifer Burnette Topic: Summer Mentorship Program (ends at 9:15) Track 5D (Grand Ballroom) – Communications & Outreach Working Group Moderators: Noel Burtt and MacKenzie Brandes Topic: Charter Finalization |
| 9:00 – 9:30 Track 5A (Bethesda Potomac) – Ontology Working Group Moderators: Mano Maurya and Srini Ramachandran Topic: Overview of C2M2 and metadata submission (8:30-9:15am) Track 5B (Rockville/Chevy Chase) – Knowledge Graph Working Group Moderators: Deanne Taylor and Jonathan Silverstein Topic: Community Updates Track 5C (Salon E) – Trainers Working Group Moderators: Allissa Dillman and Jennifer Burnette Topic: Landscape Analysis Results and Collaboratively working to address Recommendations Track 5D (Grand Ballroom) – Communications & Outreach Working Group Moderators: Noel Burtt and MacKenzie Brandes Topic: Outreach resources and Tools |
| 9:30-10:00 Track 5A (Bethesda Potomac) – Ontology Working Group Moderators: Mano Maurya and Srini Ramachandran Topic: Addressing pending topics/questions from OWG meetings (9:15-10:00) Track 5B (Rockville/Chevy Chase) – Knowledge Graph Working Group Moderators: Deanne Taylor and Jonathan Silverstein Topic: Technologies and standards; LLMs and KGs Track 5C (Salon E) – Trainers Working Group Moderators: Allissa Dillman and Jennifer Burnette Topic: None Track 5D (Grand Ballroom) – Communications & Outreach Working Group Moderators: Noel Burtt and MacKenzie Brandes Topic: Future Events |
10:00 – 10:15 – Coffee Break
| Breakout: Partnership & Science |
|---|
| 10:15 – 11:00 Track 6A (Bethesda Potomac) Biomarker (Closed Partnership Meeting) Track 6B (Rockville/Chevy Chase) Gene Set Curation (Open to all) Track 6C (Salon E) Community Visualization Hub: Collaborative Data Visualization Platform for Genomics and Single-cell data (Open to all) Topic: End User Perspectives: Join if you want to use hub in your research |
| 11:00 – 11:45 Track 6A (Bethesda Potomac) Metabotyping (Closed Partnership Meeting) Track 6B (Rockville/Chevy Chase) Gene Set Curation (Open to all) Track 6C (Salon E) Community Visualization Hub: Collaborative Data Visualization Platform for Genomics and Single-cell data (Open to all) Topic: Tool Builder Perspectives: Join if you want to incorporate the hub in your software |
| 11:45 – 1:00 (On-site) Lunch |
| ↓ Interactive Panels: Lessons & Future ↓ (Grand Ballroom) |
| 1:00 – 1:30 Bridging Communities: Integrating CFDE with External Data Networks Moderator: Jeffrey Grethe |
| 1:30 – 2:00 Sustaining the CFDE Ecosystem Moderators: Peipei Ping and Wei Wang |
| 2:00 – 2:30 Strategies to Grow the CFDE User Base Moderators: Bernard de Bono and Noel Burtt |
| 2:30 – 3:00 Townhall |
| 3:00 – 3:10 Conclusion Moderator: NIH |
| END OF DAY 2 |
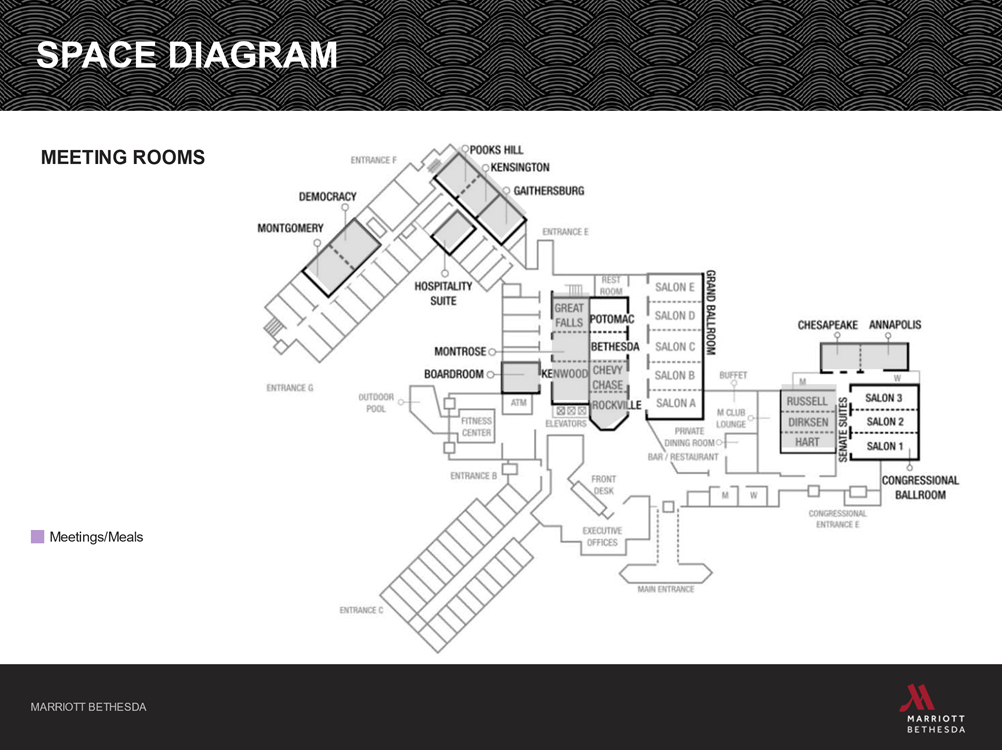
Floor Plan
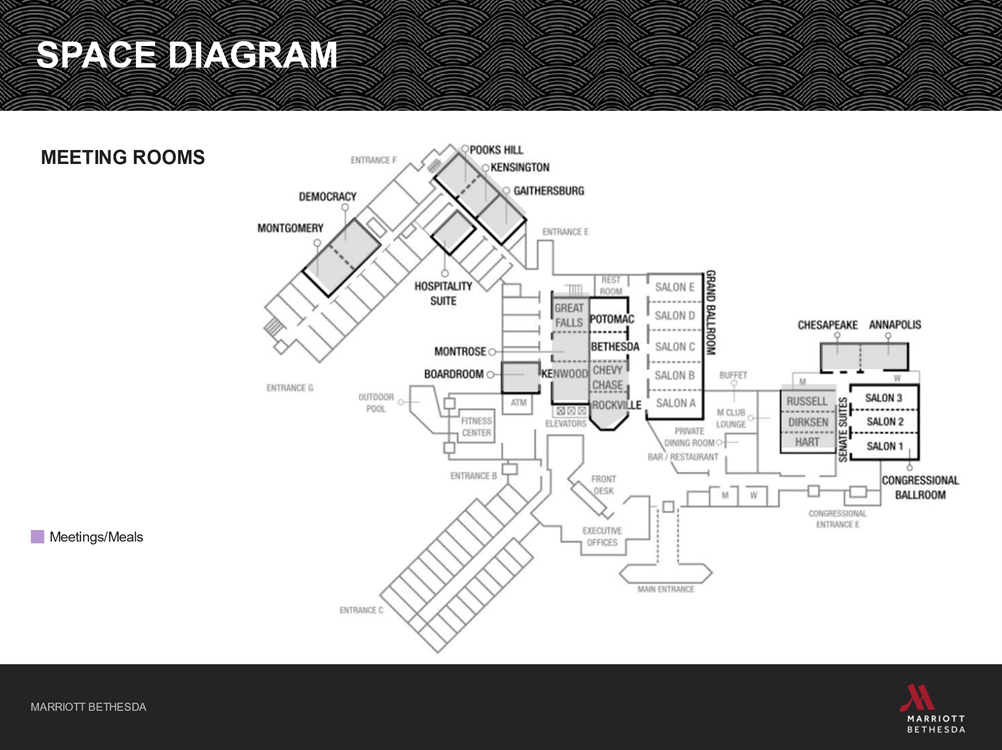
FAQ
Who can attend?
This meeting is intended for all consortium members.
Who can I contact if I have questions?
If you have any questions or concerns please contact Swathi Thaker at snthaker@uab.edu
How do I register?
Registration closed on March 1, 2025.
Will meals be provided?
Both Breakfast and Lunch will be provided onsite.
Will there be a virtual option?
This meeting is designed to be an in-person event and we highly encourage everyone to attend on site. However, if you need to attend virtually, you can use the below Zoom link:
Join Zoom Meeting
https://uab.zoom.us/j/89520549569?pwd=Lcbc5YKniOW158oyKQdox9dN6vDbtk.1
Meeting ID: 895 2054 9569
Passcode: 276611
One tap mobile
+16468769923,,89520549569# US (New York)
+13126266799,,89520549569# US (Chicago)
Where are the sessions located?

Dining Options
For those interested, the hotel offers complimentary shuttle service to Downtown Bethesda. It has stops at Medical Center Metro Station and Bethesda Downtown (Marriott Headquarters). The Shuttle runs every hour from 6:30am – 7:30pm. Pick up times from each shuttle stop is approximately 10 minutes after the posted shuttle departure time. Below is a list of restaurants that are relatively.
Below is a list of restaurants recommended by the hotel.
Italian
- Pinstripes
11920 Grand Park Ave, North Bethesda, MD 20852
(240) 630-3222
- Mama Lucia
12274 Rockville Pike M, Rockville, MD 20852
(301) 770-4894
- Chef Tony’s Amalfi
12307 Wilkins Ave, Rockville, MD 20852
(301) 770-7888
Indian
- Commonwealth Indian
11610 Old Georgetown Rd, Rockville, MD 20852
(240) 833-3055
- Om Indian
785 Rockville Pike, Rockville, MD 20852
(301) 279-7700
- Bombay Bistro
98 W Montgomery Ave, Rockville, MD 20850
(301) 762-8798
Thai
- Tara Thai
12071 Rockville Pike, Rockville, MD 20852
(301) 231-9899
- Bangkok Garden
891 Rockville Pike, Rockville, MD 20852
(301) 545-2848
- Taipei Tokyo
11510 Rockville Pike, Rockville, MD 20852
(301) 881-8388
Seafood
- Seasons 52
11414 Rockville Pike, North Bethesda, MD 208S2
(301) 984-5252
- Bethesda Crab House
4958 Bethesda Ave, Bethesda, MD 20814
(301) 652-3382
- Live Crawfish & Seafood
765 Rockville Pike Ste F-G, Rockvllle, MD 20852
(301) 978-7988
American
- Clydes Tower Oaks Lodge
2 Preserve Pkwy, Rockville, MD 20852
(301) 294-0200 - Summer House
11825 Grand Park Ave, North Bethesda, MD 20852
(301) 881-2381 - Matchbox
1699 Rockville Pike, Rockville, MD 20852
(301) 816-0369
Sushi
- Kusshi Sushi
11826 Trade St, North Bethesda, MD 20852
(240) 770-0355 - Hinode
134 Congressional Ln, Rockville, MD 20852
(301) 816-2190 - Sushi Oma
967 Rose Ave, North Bethesda, MD 20852
(301) 664-3129
Latin
- Nada
11886 Grand Park Ave, North Bethesda, MD 20852
(301) 770-4040
- La Brasa
12401 Parklawn Dr, Rockville, MD 20852
(301) 468-8850
- Marquez Pupuseria
890 Rockville Pike, Rockville, MD 20852
(240) 855-9781
Asian
- Pho Eateries
11618 Rockville Pike, Rockville, MD 20852
(240) 669-9777
- Far East Restaurant
5055 Nicholson Ln, Rockville, MD 20852
(301) 881-5552 - China Garden
11333 Woodglen Dr, Rockville, MD 20852
(301) 881-2800
Pizza
- Stella Barra
1182S Grand Park Ave, North Bethesda, MD 208S2
(301) 770-8609 - Armands
190 Halpine Rd, Rockville, MD 20852
(301) 231-5000 - MOD Pizza
12027 Rockville Pike, Rockville, MD 20852
(301) 287-4284
Vegan/Vegetarian
- Siena’s
4840 Boiling Brook Pkwy, North Bethesda, MD 208S2
(301) 770-7474 - Flower Child
1020S Old Georgetown Rd, Bethesda, MD 20814
(301) 664-4971 - PLNT Burger
113SS Woodglen Dr, Rockville, MD 208S2
(202) 933-5414
Bar
- Hank Dietle’s Tavern
11010 Rockville Pike, Rockville, MD 20852
(240) 512-3077 - Quincy’s South
11401 Woodglen Dr, Rockville, MD 20852
(240) 669-3270 - Owens Tavern
11820 Trade St, North Bethesda, MD 20852
(301) 245-1226
